-Search query
-Search result
Showing 1 - 50 of 58 items for (author: kunz & s)

EMDB-19440: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

PDB-8rqf: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

EMDB-17207: 
Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor
Method: single particle / : Greenhough LA, Liang CC, West SC

EMDB-17114: 
CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI, Maslen S, Skehel JM

EMDB-17115: 
CAND1-CUL1-RBX1-DCNL1
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI, Maslen SL, Skehel JM

EMDB-17116: 
CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI, Maslen S, Skehel JM

EMDB-17117: 
Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI, Maslen SL, Skehel JM

PDB-8or0: 
CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI

PDB-8or2: 
CAND1-CUL1-RBX1-DCNL1
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI

PDB-8or3: 
CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI

PDB-8or4: 
Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI

EMDB-17205: 
Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 3.4 A resolution
Method: single particle / : Greenhough LA, Liang CC, West SC

EMDB-17206: 
Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 2.2 A resolution
Method: single particle / : Greenhough LA, Liang CC, West SC

PDB-8ouy: 
Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 3.4 A resolution
Method: single particle / : Greenhough LA, Liang CC, West SC

PDB-8ouz: 
Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 2.2 A resolution
Method: single particle / : Greenhough LA, Liang CC, West SC

EMDB-17124: 
CAND1-CUL1-RBX1
Method: single particle / : Shaaban M, Clapperton JA, Ding S, Maeots ME, Enchev RI

EMDB-12543: 
Hantaan virus-like particle glycoprotein lattice.
Method: subtomogram averaging / : Stass R, Rissanen I, Bowden TA, Huiskonen JT

EMDB-12544: 
Hantaan virus-like particle glycoprotein spike in complex with Fab fragment HTN-Gn1.
Method: subtomogram averaging / : Stass R, Rissanen I, Bowden TA, Huiskonen JT

PDB-7nrh: 
Hantaan virus glycoprotein (Gn) in complex with Fab fragment HTN-Gn1.
Method: subtomogram averaging / : Stass R, Rissanen I, Bowden TA, Huiskonen JT

EMDB-12817: 
Chaetomium thermophilum Chl1 Helicase
Method: single particle / : Hodakova Z, Singleton MR

EMDB-11678: 
Cadherin fit into cryo-ET map
Method: subtomogram averaging / : Sikora M, Ermel UH, Seybold A, Kunz M, Calloni G, Reitz J, Vabulas RM, Hummer G, Frangakis AS

PDB-7a7d: 
Cadherin fit into cryo-ET map
Method: subtomogram averaging / : Sikora M, Ermel UH, Seybold A, Kunz M, Calloni G, Reitz J, Vabulas RM, Hummer G, Frangakis AS

EMDB-10626: 
Negative stain map of CoREST complex (LSD1:RCOR1:HDAC1)
Method: single particle / : Song Y, Fairall L, Ragan TJ, Savva CG, Saleh A, Morone N, Schwabe JWR

EMDB-10627: 
Cryo-EM map of glutaraldehye cross-linked CoREST complex (LSD1:RCOR1:HDAC1)
Method: single particle / : Song Y, Fairall L, Ragan TJ, Savva CG, Morone N, Schwabe JWR

EMDB-10628: 
Cryo EM map of BS3 crosslinked CoREST complex (LSD1:RCOR1:HDAC1)- closed form
Method: single particle / : Song Y, Fairall L, Ragan TJ, Savva CG, Morone N, Schwabe JWR

EMDB-10629: 
Cryo EM map of BS3 crosslinked CoREST complex (LSD1:RCOR1:HDAC1) - open form
Method: single particle / : Song Y, Fairall L, Ragan TJ, Savva CG, Morone N, Schwabe JWR

EMDB-10630: 
Interaction of the CoREST complex with a nucleosome with 185 bp 601 sequence DNA and a propargylamine mimic of dimethy Lys4 histone H3
Method: single particle / : Song Y, Fairall L, Wu M, Ragan TJ, Savva CG, Morone N, Cole PA, Schwabe JWR

EMDB-10051: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

EMDB-10052: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

EMDB-10053: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

EMDB-10054: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state C
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

EMDB-10055: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

EMDB-10056: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

PDB-6rxt: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

PDB-6rxu: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

PDB-6rxv: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

PDB-6rxx: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state C, Poly-Ala
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

PDB-6rxy: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

PDB-6rxz: 
Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b
Method: single particle / : Cheng J, Kellner N, Griesel S, Berninghausen O, Beckmann R, Hurt E

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel J, Wollweber F, Pfitzner A, Muehleip A, Sanchez R, Kudryashev M, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W, Daumke O
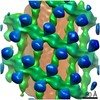
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O
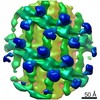
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-3753: 
Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium.
Method: single particle / : Scheffer MP

EMDB-3756: 
Sub-tomogram average of the surface adhesin (NAP) complex from Mycoplasma genitalium cells by cryo-electron tomography.
Method: subtomogram averaging / : Scheffer MP, Gonzalez-Gonzalez L, Frangakis AS
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
